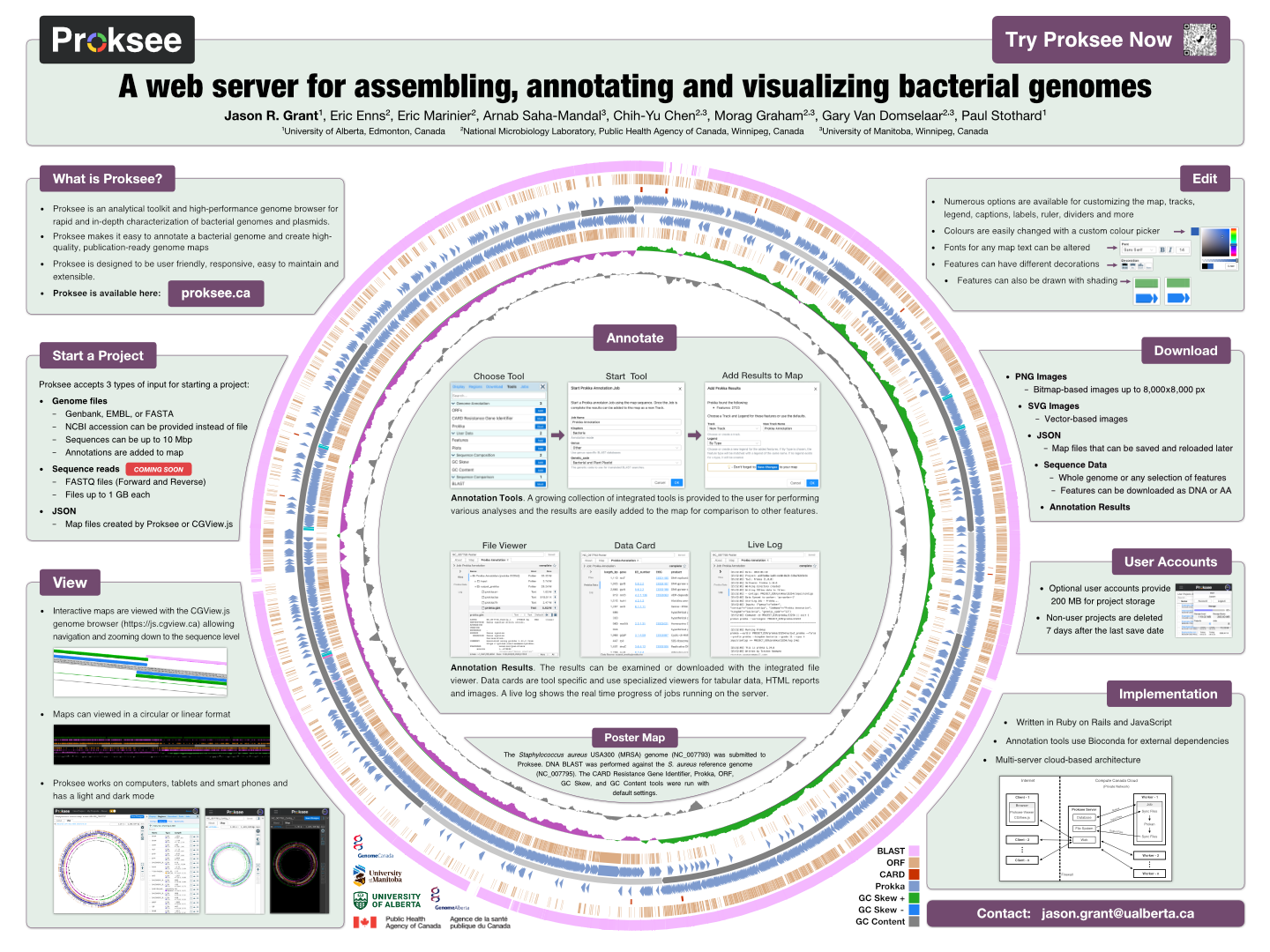
Proksee News
Proksee’s BLAST tool has been updated to make multi genome comparisons faster and easier. You can now include up to 5 BLAST subjects in a single job. Previously, you had to start a separate BLAST job for each comparison sequence. To add more subjects, click the Add File button and select another sequence (up to 5 total). Results can then be added to the map all at once, with one BLAST track per subject.
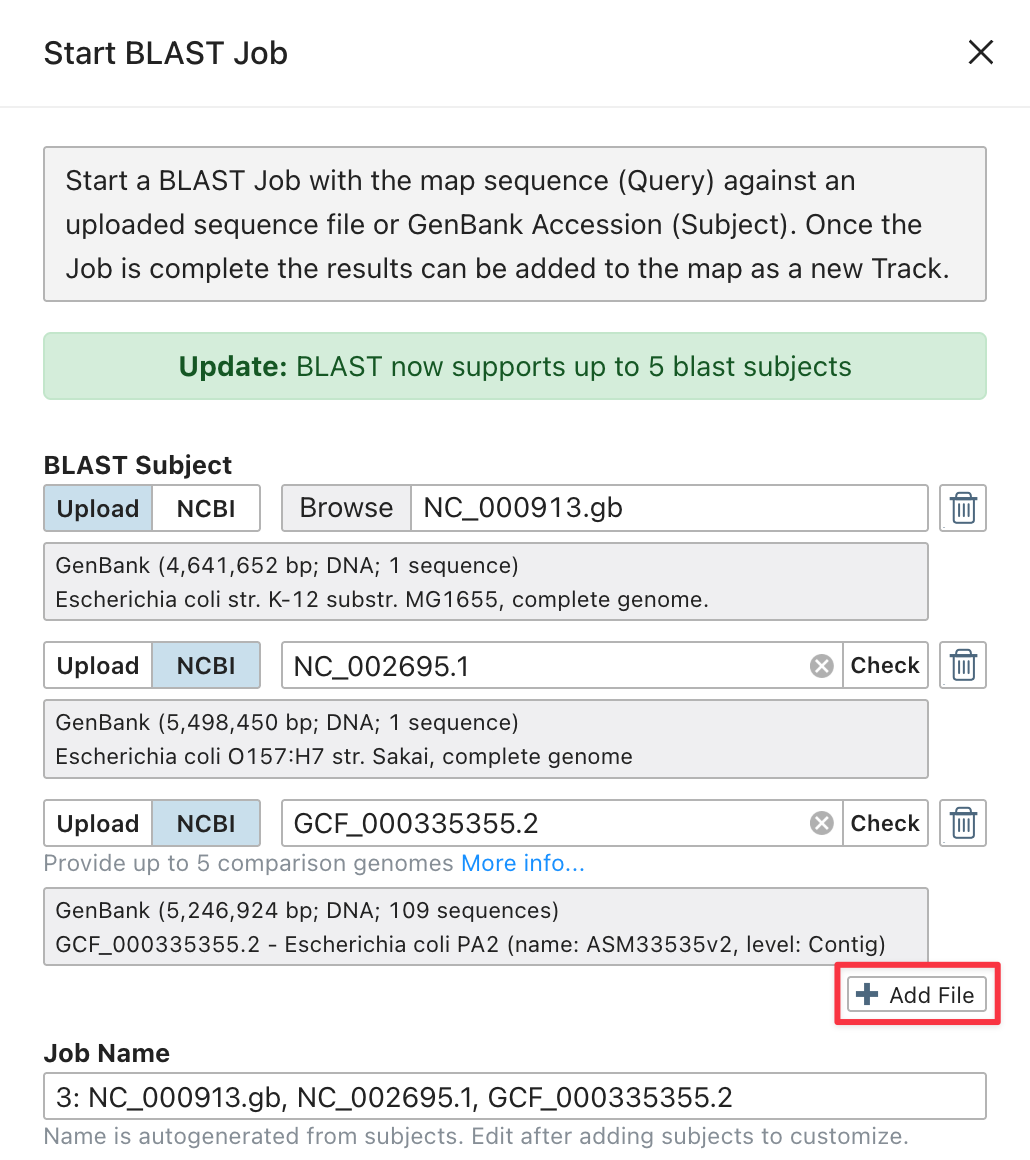
We have also reduced storage usage for BLAST jobs by removing BLAST databases and other intermediate files after the run completes.
Proksee has updated its BLAST engine (BLAST+) from 2.16 to 2.17, bringing improved performance and bug fixes.
This update corresponds to Proksee build 2026-02-24.
Update: We are now on Proksee build 2026-02-25 due to a bug introduced in the previous build.
The CARD RGI tool in Proksee has been updated to version 6.0.3, using CARD Database version 4.0.1.
For details on these releases, see the CARD website: https://card.mcmaster.ca/download
This update corresponds to Proksee build 2026-02-09.
A new data download option is now available. In addition to downloading sequences from the genome or groups of features, you can now download a specific region of the genome.
You can select a region from a specific contig or across the whole genome. A preview of the selected sequence is shown and can be copied directly to the clipboard or downloaded as a FASTA file. This simplifies extracting precise genomic regions for downstream use.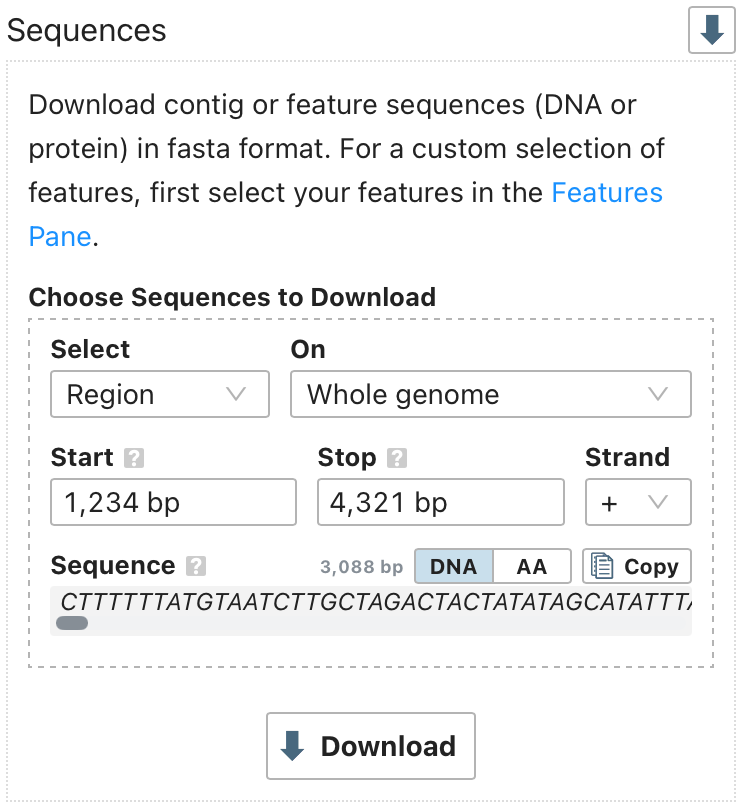
Proksee now displays a build date to help users cite the platform in publications.
Because Proksee is an evergreen web service, build dates are used instead of traditional version numbers. A build represents a stable snapshot corresponding to major features, tool updates, or significant bug fixes that may affect results.
You can find the current Proksee build date in two places:
- On the About page
- In the footer of any Proksee page
A full list of builds and associated changes is available on the new Proksee Builds (Changelog) page.
We have added a new feedback feature to Proksee. You can share comments, suggestions, and report issues using the Feedback button at the top of the page and in other places throughout Proksee. We would love to hear what is working well, what could be improved, and any ideas you have for new features or tools.
Proksee now accepts NCBI assembly accessions when creating new projects. Previously, only sequence accessions such as NC_000913 were allowed. You can now provide assembly accessions beginning with GCF or GCA, enabling direct project creation from full NCBI assemblies.
Assembly accessions are also supported in the BLAST and FastANI tools.
PNG exports are now supported at resolutions up to 16,000 × 16,000 pixels (twice the previous limit). This enables high-quality image downloads for large-format printing and detailed visual analysis. Note that extremely large sizes may still be limited by your browser or system memory.
Proksee now supports direct DNA sequence search within the genome map. Users can search for exact sequences or flexible patterns using regular expressions. To initiate a search, click the new Search Sequence button at the top of the map to open the Search sidebar.
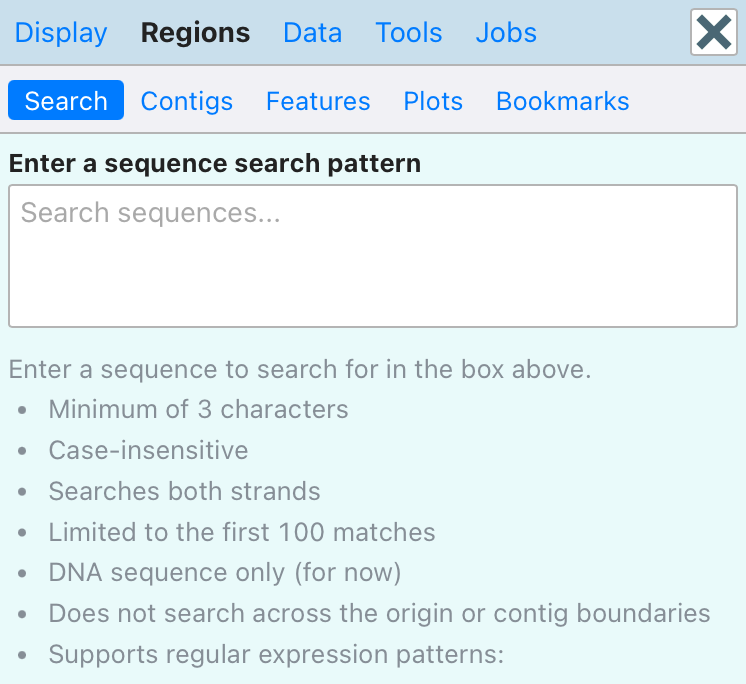
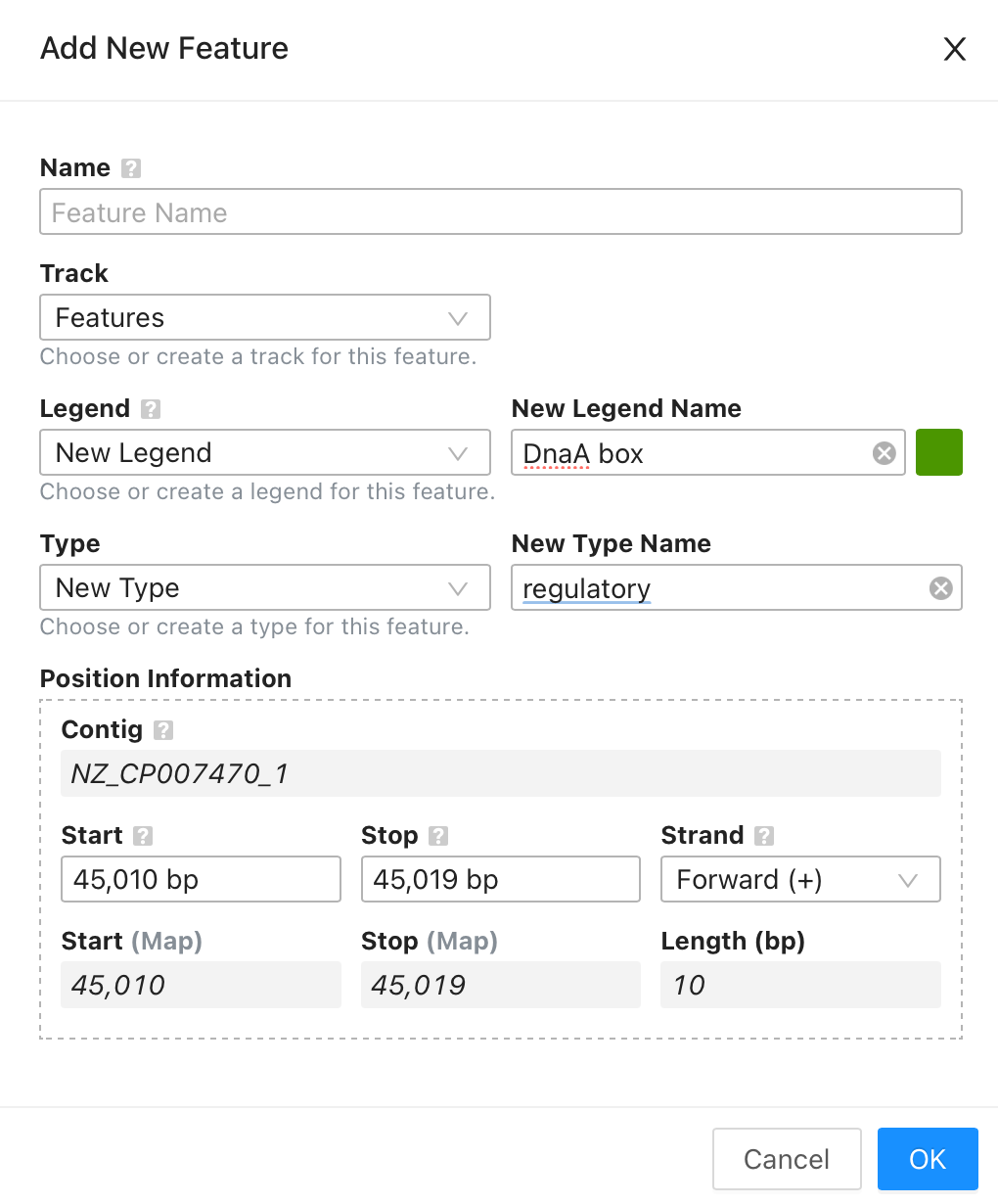
Search results can be highlighted on the map by hovering over them, and can be added as new features using the updated Add Feature dialog, accessed by clicking the button next to a result.
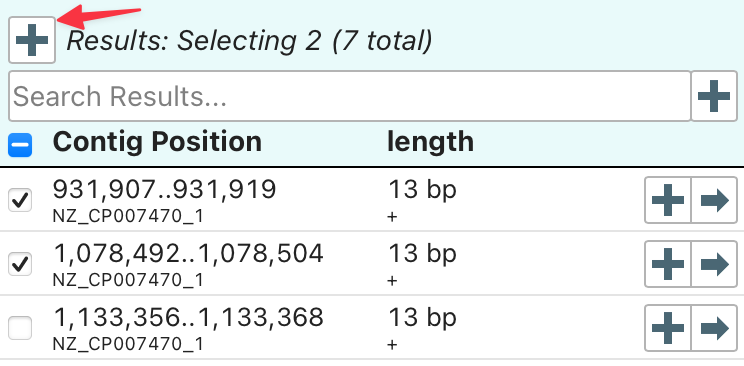
Multiple matches can be selected and added simultaneously.
Search functionality has been enhanced across all Proksee tables, including features, bookmarks, contigs, plots, tools, projects, and jobs. The most significant improvements apply to the Feature Table, which now supports searches across both standard qualifiers and tool-specific metadata.
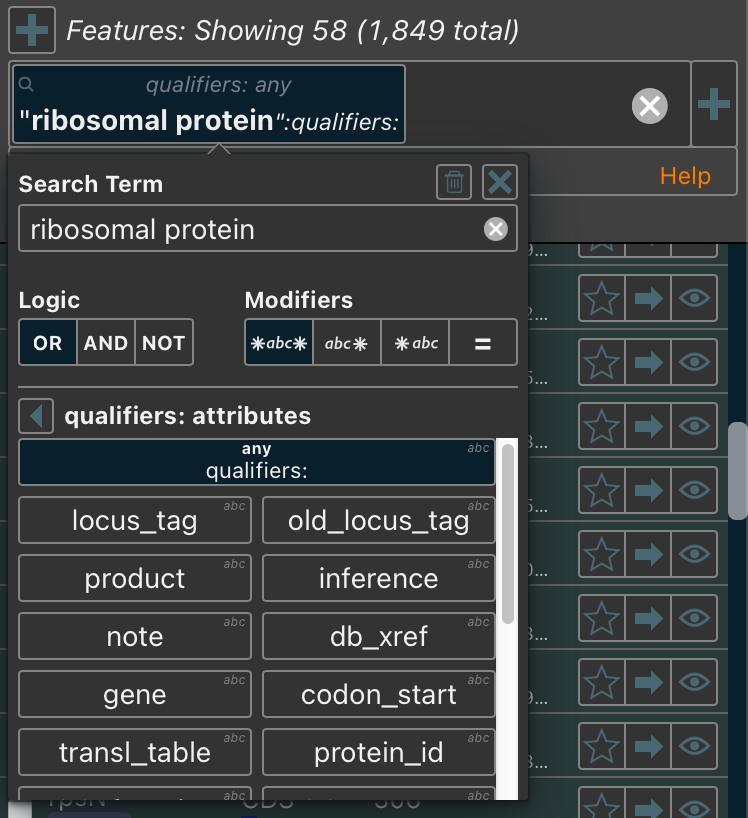
Other updates include:
- Improved query builder with a new button, located on the right side of the search bar, for quickly adding search terms
- Boolean searches (e.g. true:favorites to match favorited items) alongside numeric and string comparisons
- Saved searches, accessible below the search bar when active, can be reused across sessions on the same device.
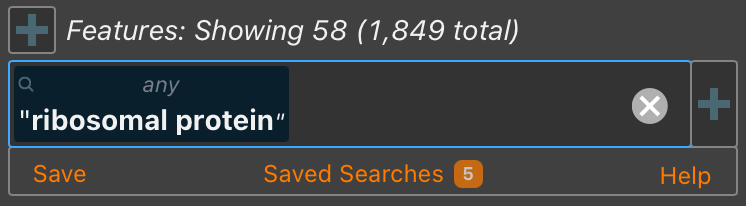
Full documentation is available in the help panel in any searchable table.
To improve communication during service interruptions and maintenance periods, we’ve launched a new Proksee Status Page.
Occasionally, tools may become unavailable or servers may experience downtime. The Status Page provides timely updates on current issues and scheduled maintenance events.
It can be accessed from the “About” page or via the “Status” link in the site footer.
When there is an active issue or scheduled maintenance:
- A banner will appear on the home page
- A real-time status icon in the top navigation bar will indicate the current system state:
: Active Issue
: Scheduled Maintenance
In the unlikely event that Proksee is fully offline, updates will be shared via our Bluesky account @proksee.ca.
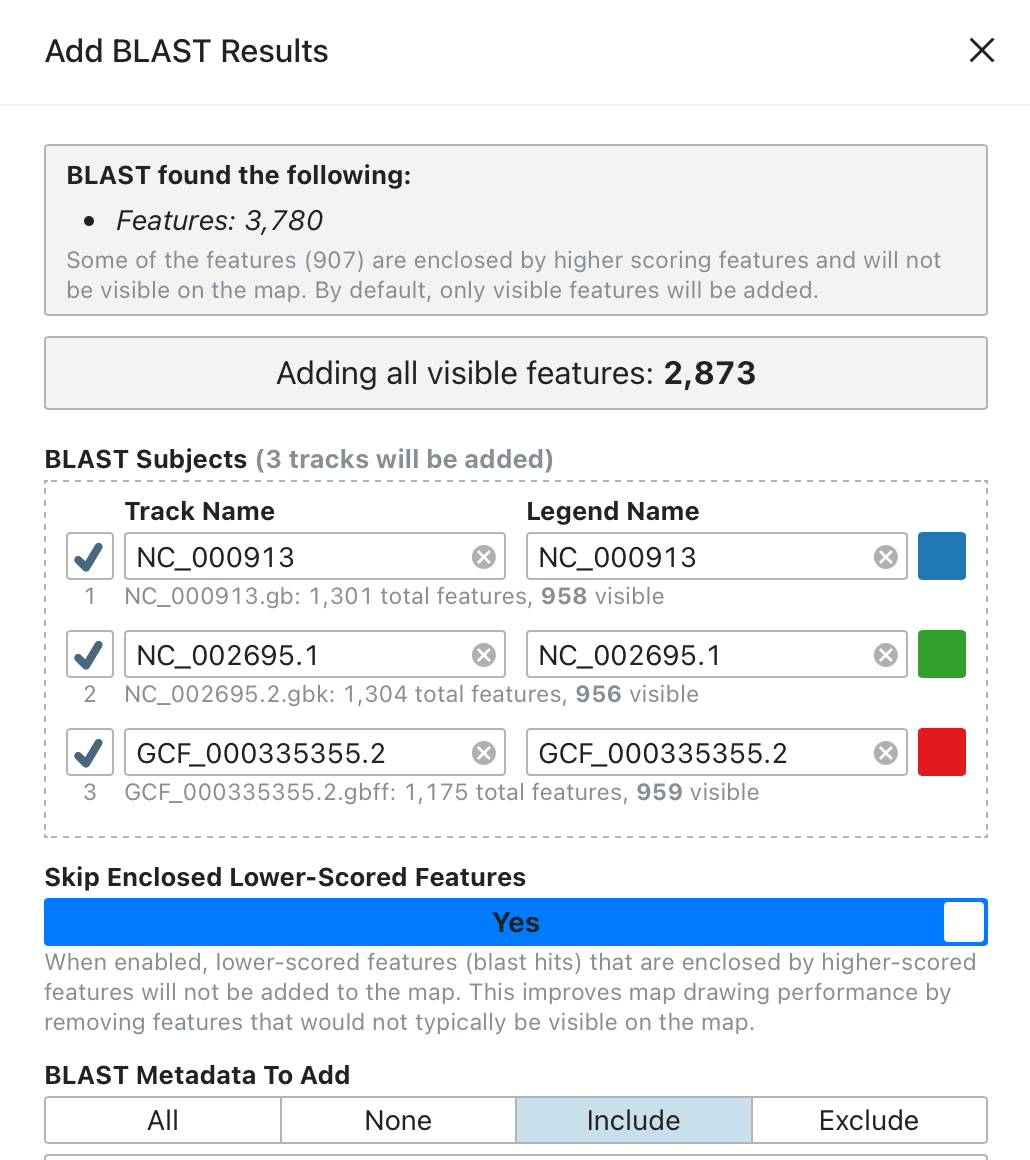
Proksee now provides enhanced control over metadata associated with BLAST and CARD results. Previously, we balanced comprehensive data inclusion with manageable file sizes, limiting metadata to the most critical fields. We've now introduced a customizable metadata selection feature to give users full control over which data to include.
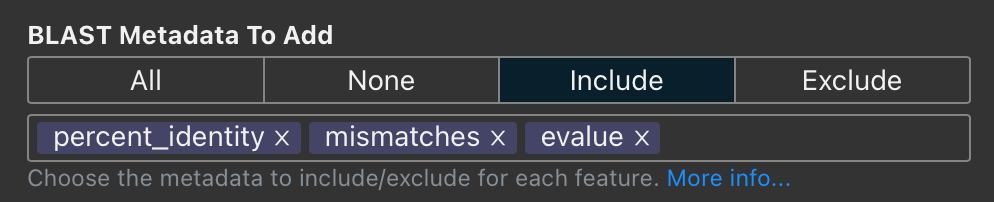
The new metadata selection interface allows users to:
- Include all metadata for maximum detail, none for minimalism, or selectively include or exclude specific fields.
- Retain their selection preferences in the browser for future use.
- Adjust metadata selections dynamically—simply re-add features with updated settings, without rerunning jobs.
Detailed Metadata Available
BLAST
- Previous: identity, mismatches, evalue, bit_score
- Current: subject, percent_identity (previously identity), mismatches, alignment_length, gap_openings, subject_start, subject_stop, evalue, bit_score
CARD
- Previous: cutoff, aro, drug_class, resistance_mechanism, amr_gene_family
- Current: cutoff, pass_bitscore, best_hit_bitscore, best_hit_aro, best_identities, aro, model_type, snps_in_best_hit_aro, other_snps, drug_class, resistance_mechanism, amr_gene_family, nudged, note, antibiotics
Selected metadata is currently displayed in feature tooltips when hovering over map elements. Later this year, Proksee will support searching based on metadata fields, enabling even more powerful data exploration.
Note: Metadata selection applies only to new BLAST and CARD jobs, as previous results do not include the expanded metadata.
Also new in this release: Proksee has updated its BLAST engine from version 2.15 to 2.16, bringing improved performance and compatibility with the latest BLAST features.
Stay tuned as we continue expanding metadata customization across more Proksee tools in the coming months.
We have enhanced the Track List Caption tool in Proksee to provide dynamic and real-time updates. Previously, the caption remained static and did not reflect changes made to the track list. With this update, the caption now automatically updates to accurately represent any additions, removals, reordering, or edits to tracks.
The Track List Caption can be added from the Tools panel or by clicking the button at the top of the Track panel:
Once a caption is created, it can also be accessed from the caption settings:
Additionally, we have introduced a new option that allows the track list numbering to start from the backbone ring:

- Previously created track list captions will remain static and do not update automatically.
- Dynamic caption text can not be edited.
- To create a static text caption with a track list you can edit, click the "Copy List" button to copy the track list to your clipboard, then create a new caption in the caption panel and paste the copied track list text.
Follow @proksee.ca on Bluesky for the latest news and updates!
We'll continue sharing updates here as well as on Bluesky.
For support, the best way to reach us is still via email: info@proksee.ca
🦋 Proksee is now on Bluesky! 🦋 #Proksee #Bacteria #Plasmids #Mitochondia #Genomics
— Proksee (@proksee.ca) February 28, 2025 at 12:58 PM
The maximum length limit for Alien Hunter has been increased from 1 Mb to 5 Mb. Additionally, we have fixed a bug that caused the optimized boundaries to be ignored when creating features.
Phigaro has been updated to 2.4.0 (from 2.3.0).
This version of Phigaro fixed a bug with gff3 and bed files formats: prophage coordinates were shifted by 1 nucleotide upstream in versions phigaro <= 2.3.0.
Proksee has changed the type for the features to use consistent GenBank feature keys.
See the CHANGELOG for a full list of changes.
A new tool has been added to Proksee:
- PHASTEST
- Detect and annotate prophage regions with PHASTEST.
- Paper: PHASTEST: Faster than PHASTER, Better than PHAST
Starting today, the Alien Hunter tool on Proksee is limited to analyzing a single sequence between 20,000 bp and 1,000,000 bp in length. This change is due to ongoing performance and stability issues that have affected its reliability. If these issues persist, the tool may be discontinued entirely. However, if performance improves, we may consider increasing the maximum sequence size in the future.
For analyzing longer genomes or multiple contigs, we recommend alternative tools in the Mobile Genetic Elements category, such as VirSorter and Phigaro. Additionally, we will soon be adding PHASTEST, a popular tool for identifying prophage regions.
Thank you for your understanding and continued support! If you have any questions or concerns, please feel free to reach out at info@proksee.ca.
Proksee now includes Snapshots, a convenient way to save and revisit genome map states and share them with others. The new Snapshot button above the map saves the current map state as a Map JSON file on the Proksee server.
A snapshot is functionally equivalent to downloading and reloading the Map (CGView) JSON from the Download Panel, with the added benefit of staying within the project. Snapshots do not include job results, and jobs within the project remain unaffected when viewing or restoring snapshots.
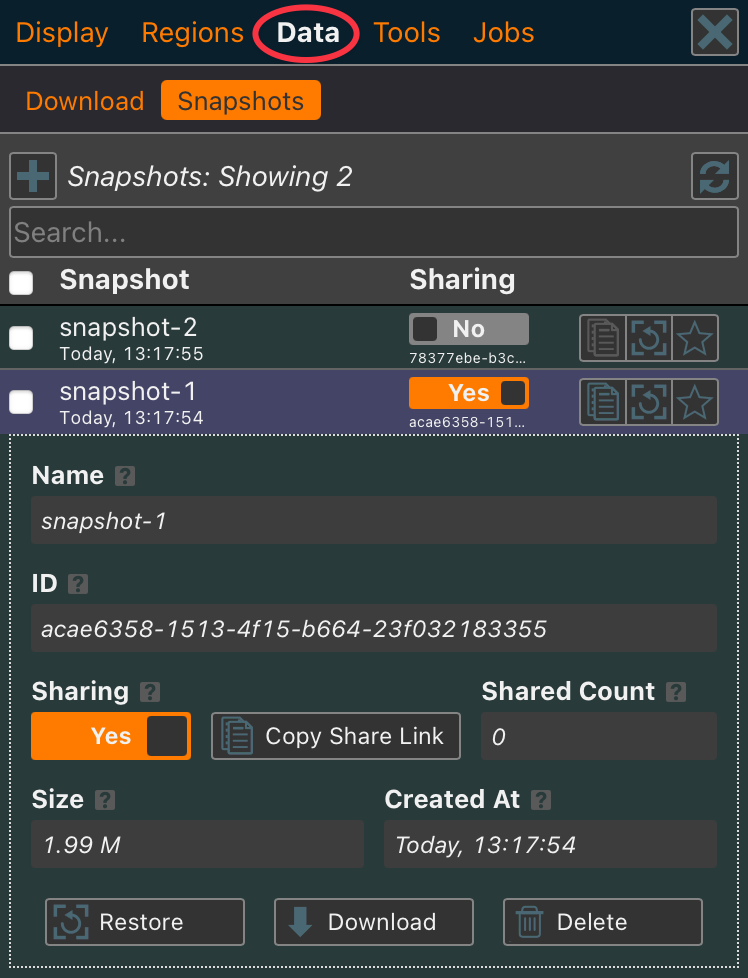
The Download tab has been replaced with a Data tab, which includes both the Download and Snapshots panels. From here, you can restore previous snapshots or enable sharing. Shared snapshots generate a link that can be provided to others. When a shared link is used, it creates a new project based on the snapshot, just as if it were created from a Map JSON file.
A Map Style is a collection of visual settings (e.g. legend colors, label fonts, etc) that can be saved from one map and applied to others via a JSON file. This is ideal for reusing customized styles across multiple maps, saving time and ensuring consistency.
The Map Style button is located in the Display Pane, to the right of the Settings tab button.
The Map Style dialog provides two main functionalities, organized into separate tabs:
- Export: Save your current map settings to a JSON file. A list of customizable settings will be displayed, allowing you to choose which ones to include in the export.
- Import: Apply saved settings from a JSON file to a new map. When importing, you can select specific settings from the list to customize the map as desired.
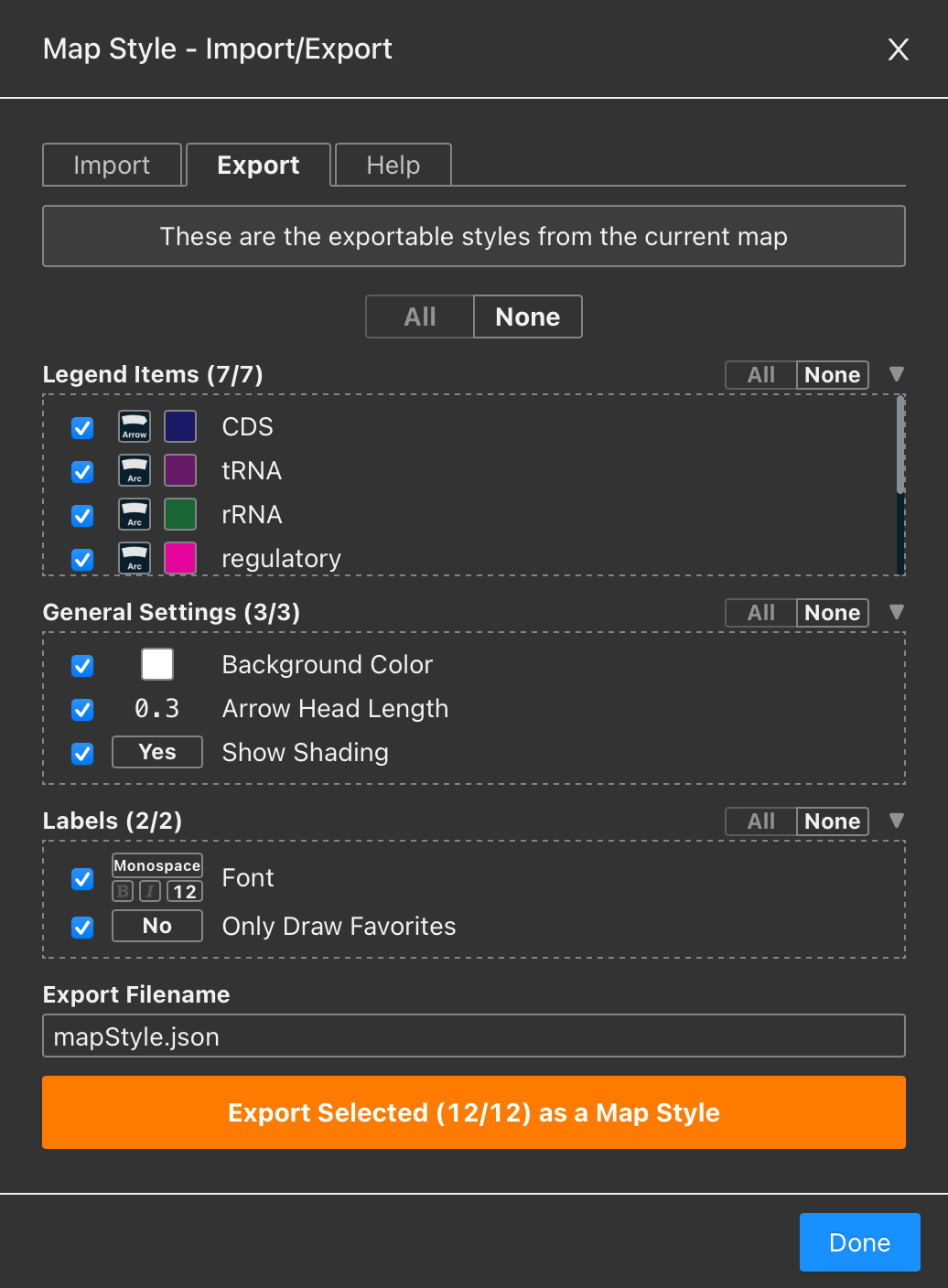
The feature detail view now includes a new Sequence section at the bottom. You can easily copy the DNA sequence for any feature. If the feature is a CDS, the amino acid sequence is also available for copying. Unless otherwise specified, the amino acid sequence is translated directly from the DNA sequence using the appropriate genetic code.

If the original GenBank or EMBL file contains a qualifier translation for the feature, and it differs from the translation derived from the DNA sequence, the original translation is retained and provided.

This is available for all features in the Features Panel:
The download sequences panel will also use the original translation if available and if the default genetic code is used.
The CGViewBuilder 2.0 parser for converting GenBank, EMBL, and FASTA files into CGView Map JSON has been completely redesigned in JavaScript, bringing several key improvements:
- Enhanced Error Handling: Now supports non-standard GenBank files generated by third-party tools (e.g., SnapGene, Geneious), minimizing parsing errors.
- Improved Performance: Genome files are processed directly in the browser, providing a faster and more streamlined workflow.
- Support for Complex Features: Features with multiple locations are no longer skipped and are displayed as joined feature segments:

- Detailed Log: After uploading a genome, a log is generated that outlines the parsing process and highlights any errors or warnings encountered during the conversion.
- New Import Settings: Access additional options via the settings button:
- Customizable Feature Names: Select which feature qualifiers to use (and their order) when choosing feature names.
- Feature Types Selection: Control which feature types to include or exclude from the map.
- Qualifier Selection: Choose which feature qualifiers to include or exclude. Qualifiers will appear in popups when hovering over features and, in the future, will be viewable, editable, and searchable in the Feature Pane.
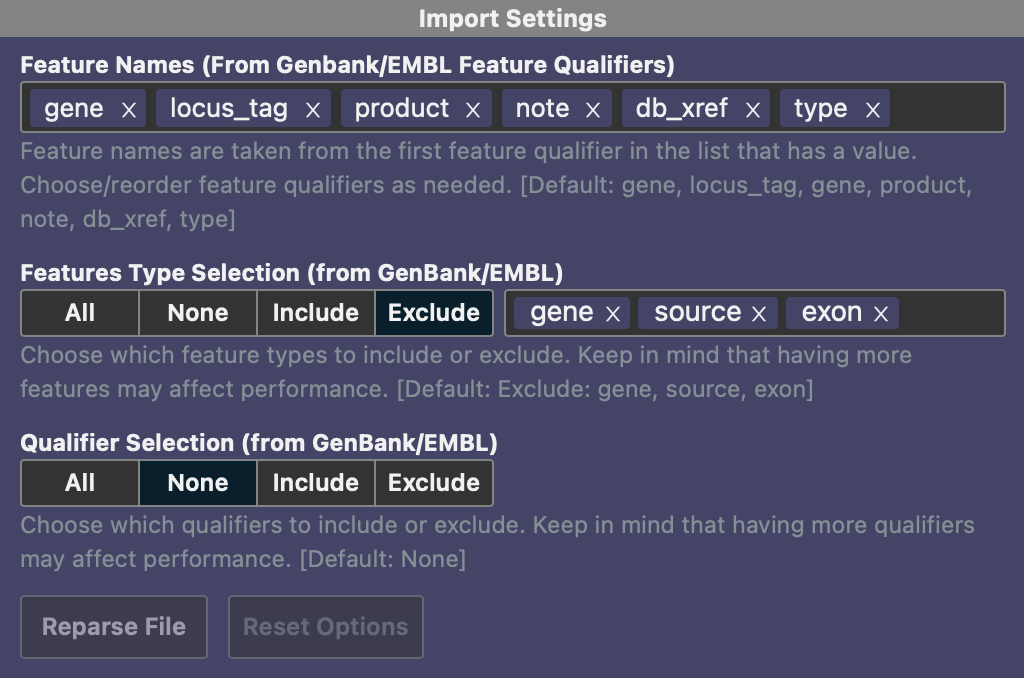
The feature tool has been fully rewritten to support a wider range of file types. In addition to CSV/TSV, it now imports features from GFF3, GTF, and BED files. The file format is automatically detected, but if the wrong format is selected, you can manually override it in Import Settings using the settings button. This is also helpful if you have a GTF, GFF3, or BED file but want to import specific columns—simply select TSV, reparse, and choose the desired columns. In the example below, only the type, start and stop positions will be imported from the provided GTF file.
A new detailed log is also available. After uploading a features file, a log is generated that outlines the parsing process and highlights any errors or warnings encountered during the conversion.
Two new options have been added to the Separate Features By setting for tracks. In addition to separating features by strand or reading frame, you can now separate them by type or legend. This update displays a new ring or lane on the map for each type or legend, with the rings/lane layers sorted so that those with the most features appear at the bottom or closer to the center of the map.
An optional center line has been added to the map view, providing a useful guide for aligning features, plots, and the sequence. The center line can be easily toggled on or off using the new button in the Format Bar. Customization options, such as color and thickness, are available in the Sidebar for further adjustments.
Tool options in the Start/Add dialogs now remember changes when rerunning tools. For example, in the BLAST tool, the default value for "Filter low complexity sequences" is Yes. If you change this to No, the tool will remember that change for the next time you run BLAST. These changes are saved in your browser's LocalStorage (similar to cookies).
At the bottom of tool Start/Add dialogs, is a new "Reset Options" button. This button resets the options to the default values. If the options are already set to the defaults the button will be disabled.
CARD RGI has been updated to 6.0.3 (from 6.0.2).
This version of CARD adds HSP subject coordinates and antibiotics to output.
The default storage quota for Proksee user accounts has increased to 500 MB (from 200 MB).
Proksee now reports raw (unadjusted) values for base composition plots. These values can be obtained by hovering over plot regions, and summary statistics (minimum, maximum, and average values) can be accessed in the Plots Panel.
The tool documentation has been updated to better describe how the GC Content and GC Skew plot values are generated and displayed.
GC Content values are calculated using a sliding window and the formula (G+C)/(Length), which leads to values between 0 and 1.
GC Skew values are calculated using a sliding window and the formula (G-C)/(G+C), which leads to values between -1 and 1.
The Y-axis range of displayed plots is automatically set to the min/max values for the plot (e.g. if the minimum score is 0.26, the Y-axis will start at 0.26).
This Y-axis adjustment allows the full plotting space to be utilized and variation to be more easily observed.
A new tool has been added to Proksee:
- Bakta
- Annotate the genome sequence with Bakta.
- Paper: Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification
A new tool has been added to Proksee:
- MITOS
- Annotate metazoan mitochondrial genomes with MITOS 2.
- Paper: Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes
A new tool has been added to Proksee:
- Track List Caption (Map Refinements)
- Creates a list of visible track rings (circular) or lanes (linear)
- This list can be added to the map as a caption or copied to the clipboard
- The tool is also accessible from the top of the Track Pane by clicking the button
A new tool has been added to Proksee:
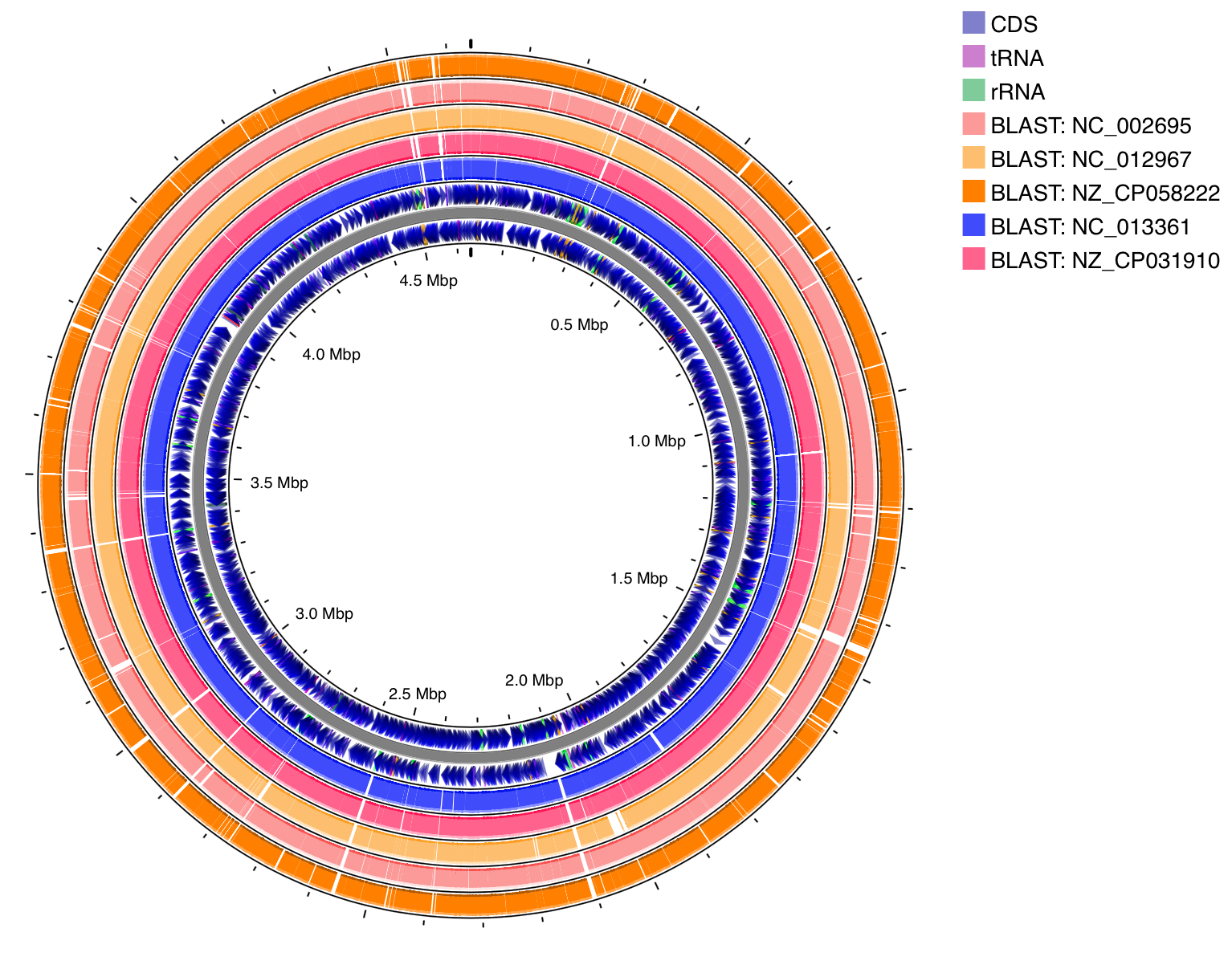
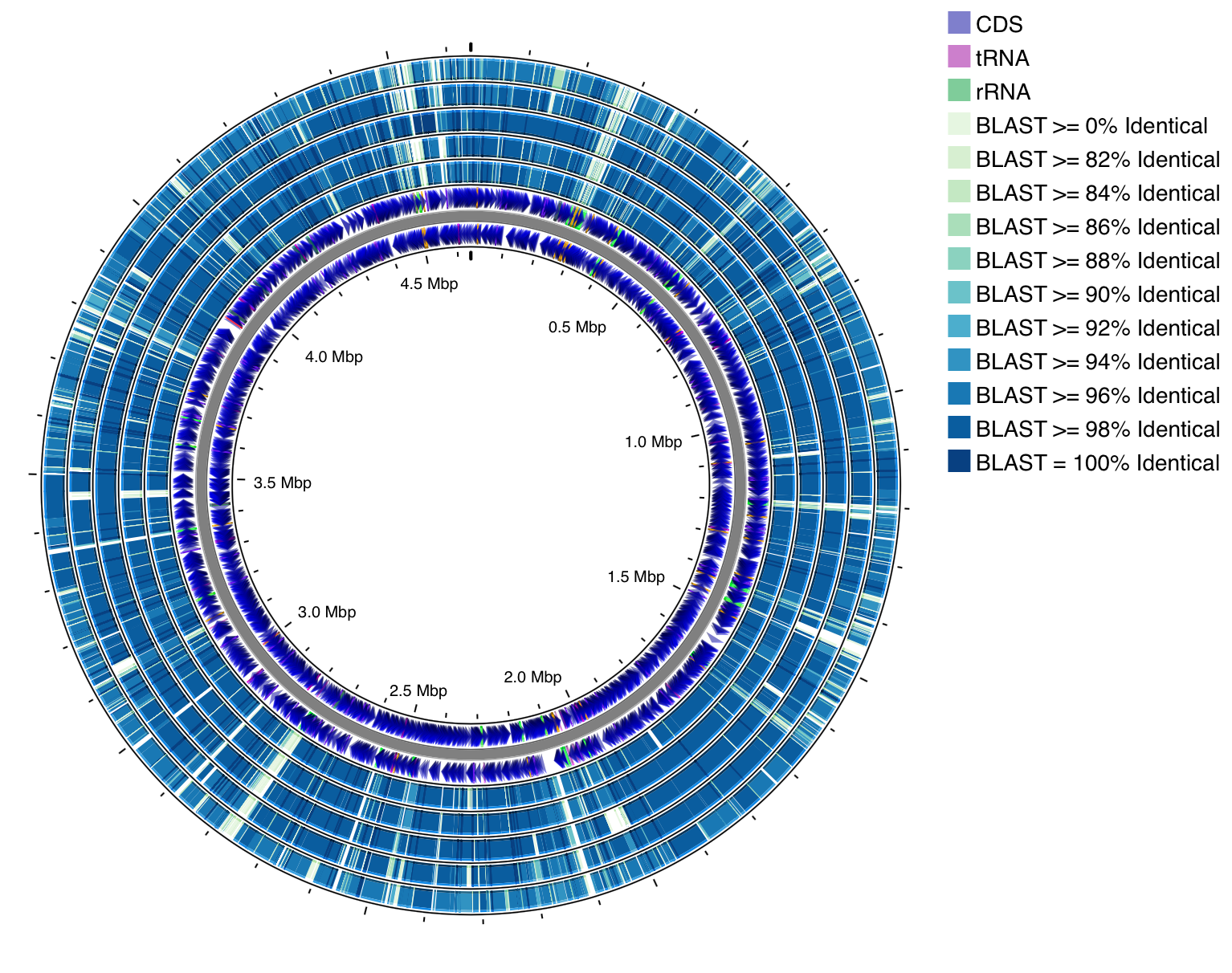
- BLAST Formatter (Map Refinements)
- Applies a customizable color scheme to the results from one or more BLAST comparisons based on their percent identity.
- Sorts BLAST tracks based on how similar they are to the map genome.
Here's an example showing E. coli str. K-12 compared against five other E. coli genomes. Before formatting (top), the hits are colored based on BLAST track (i.e. the hits from a particular job are all assigned the same color). After using BLAST Formatter (bottom), the hits are colored based on percent identity.
Within the BLAST Formatter tool you can choose the percent identity cut-offs and color scheme. A graph showing the distribution of the percent identities for the current BLAST results can be used to choose informative cut-offs.
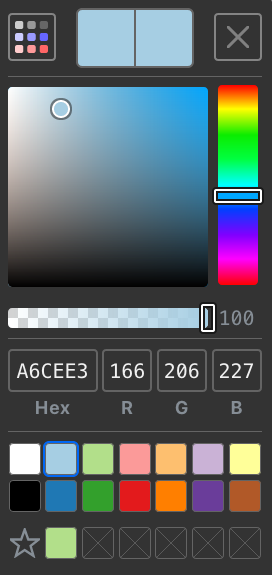
The Proksee color picker has been completely redesigned with several new features:
- Editable RGB and Hex color values.
- Swatch mode for quickly selecting predefined colors.
- Favorites for saving and using colors across maps.
More details and tips for using the color picker can be found in help.

CARD RGI has been updated to 6.0.2 (from 5.2.1).
This version of CARD removed the option --exclude_nudge and replaced it with --include_nudge for RGI main so nudged results are no longer included in default RGI main output.
New Format Bar Buttons:
Toggle Legend
Toggle Default Labels
Toggle Angled Labels
Label Notes
- Default labels work best for maps with many evenly distributed labels.
- Angled labels work best for maps with fewer labels or with labels that are clustered together.
- Both default and angled labels can be turned off to show no labels.
New Map Keyboard Shortcuts
- [↓]: Zoom In
- [↑]: Zoom Out
- [←]: Move Left/Counter-Clockwise
- [→]: Move Right/Clockwise
- [ . ]: Reset Map
- [ / ]: Toggle map format between linear and circular
If you use Proksee in your research, please cite our new paper:
Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C, Graham M, Van Domselaar G, and Stothard P
Proksee: in-depth characterization and visualization of bacterial genomes
Nucleic Acids Ressearch, 2023, gkad326, https://doi.org/10.1093/nar/gkad326
Two new tools have been added to Proksee:
- VirSorter
- Detect dsDNA and ssDNA virus genomes (phage).
- Paper: VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses
- FastANI
- Fast alignment-free computation of whole-genome Average Nucleotide Identity (ANI).
- Paper: High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries
A new tool has been added to Proksee:
- mobileOG-db
- Find mobile genetic elements (MGEs) in your sequence grouped into 5 major categories: integration/excision, replication/recombination/repair, transfer, stability/transfer/defense, and phage-specific processes.
- Paper: mobileOG-db: a Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements
Proksee can now assembly your raw sequence reads into an assembled genome. Proksee Assemble is undergoing beta testing. To try it out click on the Reads tab on the New Project page.
If you have any problems or suggestions, let us know at Assembly Feedback.
A new tool has been added to Proksee:
- Alien Hunter
- Predicts putative Horizontal Gene Transfer (HGT) events with the implementation of Interpolated Variable Order Motifs (IVOMs)
- Paper: Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands
Two new tools have been added to Proksee:
- CRISPR/Cas Finder
- Search for CRISPR Arrays and their associated (Cas) proteins.
- Paper: CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins
- pLannotate
- Annotate engineered plasmids. Sequences must be less 50,000 bp.
- Paper: pLannotate: engineered plasmid annotation
A new Help page has been added and can be accessed from the Navigation bar.
In addition, help icons![]() can be found throughout Proksee. Hovering over the icon will display a popup showing information pulled from the help page. Clicking on the help icon will open a new tab and take you directly to appropriate help section.
can be found throughout Proksee. Hovering over the icon will display a popup showing information pulled from the help page. Clicking on the help icon will open a new tab and take you directly to appropriate help section.
Maps can now be downloaded as SVG images:
Proksee was presented as a poster at the BioNet 2022 Conference.